Specifying Desired Nuclei to Construct a Network¶
The Library class in pynucastro provides a high level interface for reading files containing one or more Reaclib rates and then filtering these rates based on user-specified criteria for the nuclei involved in the reactions. We can then use the resulting rates to build a network.
This example uses a Reaclib snapshot downloaded from:
https://groups.nscl.msu.edu/jina/reaclib/db/library.php?action=viewsnapshots.
Reading a rate snapshot¶
The Library class will look for the library file in the working directory or in the pynucastro/library subdirectory of the pynucastro package.
When the constructor is supplied a file name, pynucastro will read the contents of this file and interpret them as Reaclib rates in either the Reaclib 1 or 2 formats. The Library then stores the rates from the file as Rate objects.
[1]:
%matplotlib inline
import pynucastro as pyna
[2]:
library_file = '20180201ReaclibV2.22'
[3]:
mylibrary = pyna.rates.Library(library_file)
Specifying Desired Nuclei¶
This example constructs a CNO network like the one constructed from a set of Reaclib rate files in the “pynucastro usage examples” section of this documentation.
This time, however, we will specify the nuclei we want in the network and allow the Library class to find all the rates linking only nuclei in the set we specified.
We can specify these nuclei by their abbreviations in the form, e.g. “he4”:
[4]:
all_nuclei = ["p", "he4", "c12", "n13", "c13", "o14", "n14", "o15", "n15"]
Now we use the Library.linking_nuclei() function to return a smaller Library object containing only the rates that link these nuclei.
We can pass with_reverse=False to restrict linking_nuclei to only include forward rates from the Reaclib library, as pynucastro does not yet implement partition functions for reverse rates.
[5]:
cno_library = mylibrary.linking_nuclei(all_nuclei, with_reverse=False)
Now we can create a network (PythonNetwork or StarKillerNetwork) as:
[6]:
cno_network = pyna.networks.PythonNetwork(libraries=cno_library)
In the above, we construct a network from a Library object by passing the Library object to the libraries argument of the network constructor. To construct a network from multiple libraries, the libraries argument can also take a list of Library objects.
We can show the structure of the network by plotting a network diagram.
[7]:
cno_network.plot()
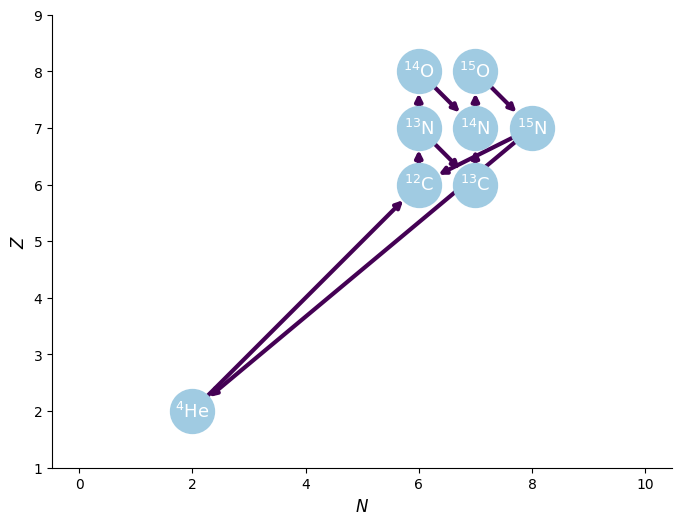
Note that the above network also includes the triple-alpha rate from Reaclib.
If we wanted to generate the python code to calculate the right-hand side we could next do:
[8]:
cno_network.write_network('network_module.py')
And we could run this together with the burn driver program in examples/burn.py