Checking Your Network#
Selecting which rates to include and which to exclude from your network is a bit of an art. pynucastro has a few tools to help check what you might be missing.
import pynucastro as pyna
RateCollection validate method#
Let’s start by trying to create a network for carbon burning.
To start, let’s pick the nuclei \(\alpha\), \({}^{12}\mathrm{C}\) and \({}^{20}\mathrm{Ne}\). We’ll explicitly read in the ReacLib library and select the rates, since we will need the library later—otherwise we could have used network_helper.
reaclib_library = pyna.ReacLibLibrary()
nuclei = ["he4", "c12", "ne20"]
cburn_library = reaclib_library.linking_nuclei(nuclei)
Now we’ll make a RateCollection (any of the other network types is fine too).
rc = pyna.RateCollection(libraries=[cburn_library])
rc
C12 ⟶ 3 He4
C12 + C12 ⟶ He4 + Ne20
Ne20 + He4 ⟶ C12 + C12
3 He4 ⟶ C12 + 𝛾
Now, since we are primarily interested in \({}^{12}\mathrm{C} + {}^{12}\mathrm{C}\), let’s make sure we are not missing any other reactions that have the same reactants. The validate method will do this, by comparing the rates we have selected to another library.
rc.validate(reaclib_library)
validation: Ne20 produced in C12 + C12 ⟶ He4 + Ne20 never consumed.
validation: missing 3 He4 ⟶ p + B11 as alternative to 3 He4 ⟶ C12 + 𝛾 (Q = -8.682 MeV).
validation: missing 3 He4 ⟶ n + C11 as alternative to 3 He4 ⟶ C12 + 𝛾 (Q = -11.4466 MeV).
validation: missing C12 + C12 ⟶ p + Na23 as alternative to C12 + C12 ⟶ He4 + Ne20 (Q = 2.242 MeV).
validation: missing C12 + C12 ⟶ n + Mg23 as alternative to C12 + C12 ⟶ He4 + Ne20 (Q = -2.598 MeV).
False
This tells us that we are missing 2 branches of the \({}^{12}\mathrm{C} + {}^{12}\mathrm{C}\) reaction. ReacLib already scales the rates based on the branching of the products, so we should try to include these other branches.
Note
By default, validate() only checks forward rates.
To address these issues, we need to include the additional nuclei. In particular, the branch that makes \({}^{23}\mathrm{Na}\) is likely important (the rate making \({}^{23}\mathrm{Mg}\) is endothermic, so less likely).
nuclei += ["p", "na23"]
cburn_library = reaclib_library.linking_nuclei(nuclei)
rc = pyna.RateCollection(libraries=[cburn_library])
rc
C12 ⟶ 3 He4
C12 + C12 ⟶ p + Na23
C12 + C12 ⟶ He4 + Ne20
Ne20 + He4 ⟶ p + Na23
Ne20 + He4 ⟶ C12 + C12
Na23 + p ⟶ He4 + Ne20
Na23 + p ⟶ C12 + C12
3 He4 ⟶ C12 + 𝛾
rc.validate(reaclib_library)
validation: Ne20 produced in C12 + C12 ⟶ He4 + Ne20 never consumed.
validation: Ne20 produced in Na23 + p ⟶ He4 + Ne20 never consumed.
validation: missing 3 He4 ⟶ p + B11 as alternative to 3 He4 ⟶ C12 + 𝛾 (Q = -8.682 MeV).
validation: missing 3 He4 ⟶ n + C11 as alternative to 3 He4 ⟶ C12 + 𝛾 (Q = -11.4466 MeV).
validation: missing C12 + C12 ⟶ n + Mg23 as alternative to C12 + C12 ⟶ He4 + Ne20 (Q = -2.598 MeV).
validation: missing C12 + C12 ⟶ n + Mg23 as alternative to C12 + C12 ⟶ p + Na23 (Q = -2.598 MeV).
validation: missing Na23 + p ⟶ n + Mg23 as alternative to Na23 + p ⟶ He4 + Ne20 (Q = -4.839 MeV).
validation: missing Na23 + p ⟶ Mg24 + 𝛾 as alternative to Na23 + p ⟶ He4 + Ne20 (Q = 11.6927 MeV).
False
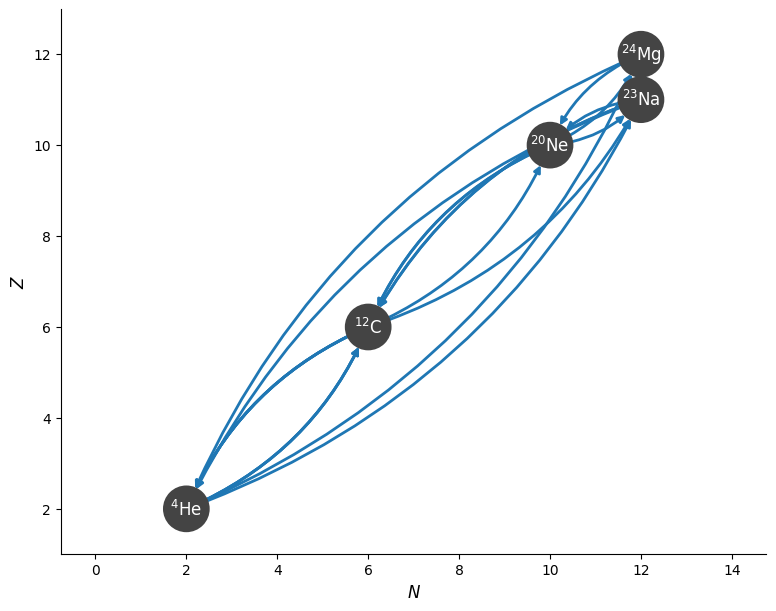
Now, looking at what is missing, we probably want to include \({}^{24}\mathrm{Mg}\) as an endpoint for carbon burning.
nuclei += ["mg24"]
cburn_library = reaclib_library.linking_nuclei(nuclei)
rc = pyna.RateCollection(libraries=[cburn_library])
rc
Mg24 ⟶ p + Na23
Mg24 ⟶ He4 + Ne20
C12 ⟶ 3 He4
Ne20 + He4 ⟶ Mg24 + 𝛾
Na23 + p ⟶ Mg24 + 𝛾
C12 + C12 ⟶ p + Na23
C12 + C12 ⟶ He4 + Ne20
Ne20 + He4 ⟶ p + Na23
Ne20 + He4 ⟶ C12 + C12
Na23 + p ⟶ He4 + Ne20
Na23 + p ⟶ C12 + C12
3 He4 ⟶ C12 + 𝛾
rc.validate(reaclib_library)
validation: Mg24 produced in Ne20 + He4 ⟶ Mg24 + 𝛾 never consumed.
validation: Mg24 produced in Na23 + p ⟶ Mg24 + 𝛾 never consumed.
validation: missing 3 He4 ⟶ p + B11 as alternative to 3 He4 ⟶ C12 + 𝛾 (Q = -8.682 MeV).
validation: missing 3 He4 ⟶ n + C11 as alternative to 3 He4 ⟶ C12 + 𝛾 (Q = -11.4466 MeV).
validation: missing C12 + C12 ⟶ n + Mg23 as alternative to C12 + C12 ⟶ He4 + Ne20 (Q = -2.598 MeV).
validation: missing C12 + C12 ⟶ n + Mg23 as alternative to C12 + C12 ⟶ p + Na23 (Q = -2.598 MeV).
validation: missing Ne20 + He4 ⟶ n + Mg23 as alternative to Ne20 + He4 ⟶ Mg24 + 𝛾 (Q = -7.21457 MeV).
validation: missing Na23 + p ⟶ n + Mg23 as alternative to Na23 + p ⟶ He4 + Ne20 (Q = -4.839 MeV).
validation: missing Na23 + p ⟶ n + Mg23 as alternative to Na23 + p ⟶ Mg24 + 𝛾 (Q = -4.839 MeV).
False
This now seems reasonable. The reactions that are missing are all endothermic.
fig = rc.plot(curved_edges=True)