Modifying Rates#
Sometimes we want to change the nuclei involved in rates to simplify our network. Currently, pynucastro supports changing the products. Here’s an example.
Note
A newer way to accomplish this is via the ModifiedRate class. In the future, the modify_products method
shown here may be removed.
import pynucastro as pyna
reaclib_library = pyna.ReacLibLibrary()
We want to model \({}^{12}\mathrm{C} + {}^{12}\mathrm{C}\) reactions. There are 3 rates involved.
filter = pyna.RateFilter(reactants=["c12", "c12"])
mylib = reaclib_library.filter(filter)
mylib
C12 + C12 ⟶ He4 + Ne20 [Q = 4.62 MeV] (C12 + C12 --> He4 + Ne20 <reaclib_cf88>)
C12 + C12 ⟶ p + Na23 [Q = 2.24 MeV] (C12 + C12 --> p + Na23 <reaclib_cf88>)
C12 + C12 ⟶ n + Mg23 [Q = -2.60 MeV] (C12 + C12 --> n + Mg23 <reaclib_cf88>)
The rate \({}^{12}\mathrm{C}({}^{12}\mathrm{C},n){}^{23}\mathrm{Mg}\) is quickly followed by \({}^{23}\mathrm{Mg}(n,\gamma){}^{24}\mathrm{Mg}\), so we want to modify that rate sequence to just be \({}^{12}\mathrm{C}({}^{12}\mathrm{C},\gamma){}^{24}\mathrm{Mg}\)
r = mylib.get_rate_by_name("c12(c12,n)mg23")
r
C12 + C12 ⟶ n + Mg23
This has the Q value:
r.Q
-2.598
Now we modify it
r.modify_products("mg24")
r
C12 + C12 ⟶ Mg24 + 𝛾
and we see that the Q value has been updated to reflect the new endpoint
r.Q
13.933577999999976
Now let’s build a network that includes the nuclei involved in our carbon burning. We’ll start by leaving off the \({}^{23}\mathrm{Mg}\)
Caution
We we get the rate object from the library via get_rate_by_name (or any similar function), we are getting
a reference to the rate in the library, not a copy. This means that our modification is reflected back in the
original library in memory. Care needs to be taken (e.g. by making a copy or rereading the library) if you want
to access the original rate.
Using our modified rate#
mylib2 = reaclib_library.linking_nuclei(["p", "he4", "c12", "o16", "ne20", "na23", "mg24"])
Now we add in our modified rate
mylib2 += pyna.Library(rates=[r])
mylib2
3 He4 ⟶ C12 + 𝛾 [Q = 7.28 MeV] (3 He4 --> C12 <reaclib_fy05>)
C12 + He4 ⟶ O16 + 𝛾 [Q = 7.16 MeV] (C12 + He4 --> O16 <reaclib_nac2>)
C12 + C12 ⟶ He4 + Ne20 [Q = 4.62 MeV] (C12 + C12 --> He4 + Ne20 <reaclib_cf88>)
C12 + C12 ⟶ p + Na23 [Q = 2.24 MeV] (C12 + C12 --> p + Na23 <reaclib_cf88>)
C12 + C12 ⟶ Mg24 + 𝛾 [Q = 13.93 MeV] (C12 + C12 --> Mg24 <reaclib_cf88>)
O16 + He4 ⟶ Ne20 + 𝛾 [Q = 4.73 MeV] (O16 + He4 --> Ne20 <reaclib_co10>)
O16 + C12 ⟶ He4 + Mg24 [Q = 6.77 MeV] (O16 + C12 --> He4 + Mg24 <reaclib_cf88>)
Ne20 + He4 ⟶ Mg24 + 𝛾 [Q = 9.32 MeV] (Ne20 + He4 --> Mg24 <reaclib_il10>)
Na23 + p ⟶ He4 + Ne20 [Q = 2.38 MeV] (Na23 + p --> He4 + Ne20 <reaclib_il10>)
Na23 + p ⟶ Mg24 + 𝛾 [Q = 11.69 MeV] (Na23 + p --> Mg24 <reaclib_il10>)
C12 ⟶ 3 He4 [Q = -7.28 MeV] (C12 --> 3 He4 <reaclib_fy05>)
O16 ⟶ He4 + C12 [Q = -7.16 MeV] (O16 --> He4 + C12 <reaclib_nac2>)
Ne20 + He4 ⟶ C12 + C12 [Q = -4.62 MeV] (Ne20 + He4 --> C12 + C12 <reaclib_cf88>)
Ne20 + He4 ⟶ p + Na23 [Q = -2.38 MeV] (Ne20 + He4 --> p + Na23 <reaclib_il10>)
Ne20 ⟶ He4 + O16 [Q = -4.73 MeV] (Ne20 --> He4 + O16 <reaclib_co10>)
Na23 + p ⟶ C12 + C12 [Q = -2.24 MeV] (Na23 + p --> C12 + C12 <reaclib_cf88>)
Mg24 + He4 ⟶ C12 + O16 [Q = -6.77 MeV] (Mg24 + He4 --> C12 + O16 <reaclib_cf88>)
Mg24 ⟶ He4 + Ne20 [Q = -9.32 MeV] (Mg24 --> He4 + Ne20 <reaclib_il10>)
Mg24 ⟶ p + Na23 [Q = -11.69 MeV] (Mg24 --> p + Na23 <reaclib_il10>)
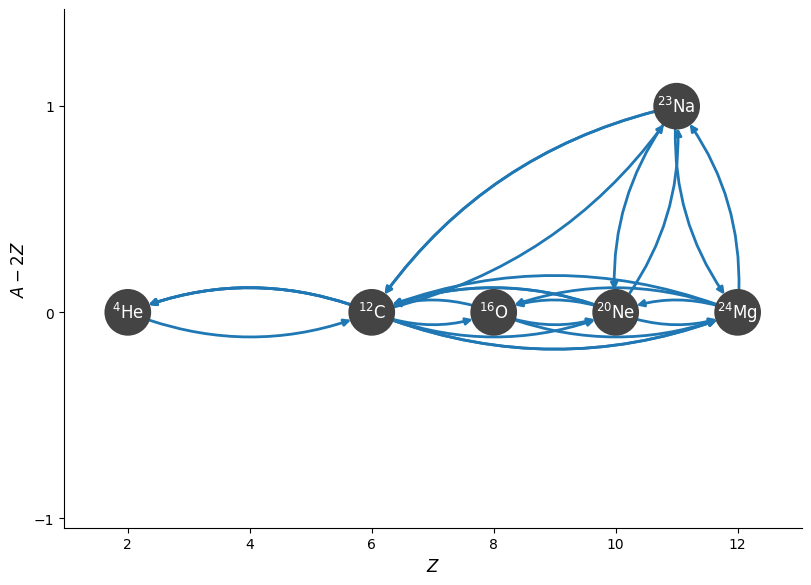
rc = pyna.RateCollection(libraries=[mylib2])
fig = rc.plot(rotated=True, curved_edges=True, hide_xalpha=True)