Computing the Screened Triple-alpha rate#
We want to evaluate the 3-\(\alpha\) rate with screening to understand how temperature sensitive it is. We’ll do this 2 different ways (but both will get the same answer).
import math
import pynucastro as pyna
We’ll define the thermodynamics first – a composition of just He and C
temp = 1.e8
dens = 1.e6
he4 = pyna.Nucleus("he4")
c12 = pyna.Nucleus("c12")
nuclei = [he4, c12]
comp = pyna.Composition(nuclei)
comp.X[he4] = 0.9
comp.X[c12] = 0.1
fig = comp.plot()
Manual method#
Here well read in this rate and compute the screening factors ourselves and then build up the full form of the rate
rl = pyna.ReacLibLibrary()
triple_alpha = rl.get_rate_by_name("a(aa,)c12")
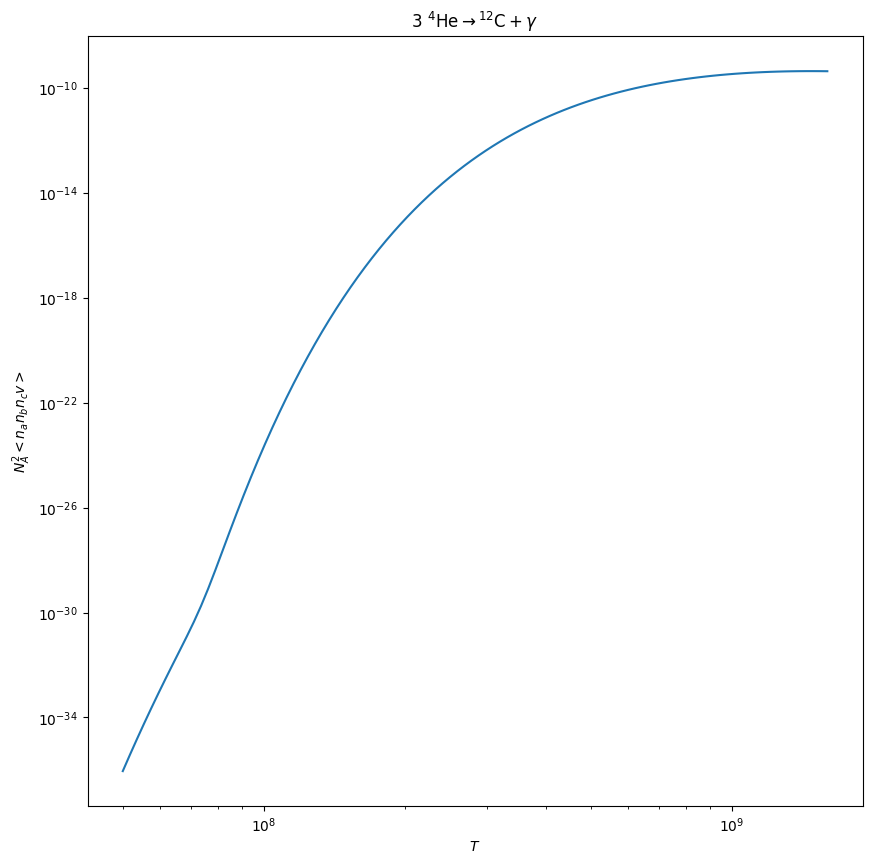
Here’s the temperature sensitivity of just the 3-\(\alpha\) rate
fig = triple_alpha.plot(Tmin=5.e7)
To compute the screening, we first need to get the plasma state
plasma = pyna.make_plasma_state(temp, dens, comp.get_molar())
Now, the 3-\(\alpha\) rate requires 2 separate screening terms, first \(\alpha + \alpha\) and then \(\alpha + {}^8\mathrm{Be}\)
scn_fac1 = pyna.make_screen_factors(he4, he4)
log_scn1 = pyna.screening.screen5(plasma, scn_fac1)
scn_fac2 = pyna.make_screen_factors(he4, pyna.Nucleus("be8"))
log_scn2 = pyna.screening.screen5(plasma, scn_fac2)
scn = math.exp(log_scn1 + log_scn2)
print(f"{scn:20.10g}")
5.802207275
This shows that our our thermodynamic conditions, screening speeds up the rate by almost \(6\times\).
Now, the total rate is:
where \(f\) is the screening factor and the \(3!\) is because there are 3 identical particles appearing in the reation.
We get \(N_A^2 \langle \sigma v \rangle\) by evaluating the temperature sensitivity of the rate from ReacLib.
r = scn * dens**2 / 6.0 * comp.get_molar()[he4]**3 * triple_alpha.eval(temp)
print(f"{r:20.10g}")
2.247437735e-14
This is the rate in units of \(\mathrm{cm}^{-3}~\mathrm{s}^{-1}\)
Computing it via a RateCollection#
We can build a simple RateCollection containing only this rate
rc = pyna.RateCollection(rates=[triple_alpha])
fig = rc.plot()
The evaluate_rates() method will take the thermodynamic state and screening function (optionally) and compute the value of each of the rates in the network.
rc.evaluate_rates(dens, temp, comp, screen_func=pyna.screening.screen5)
{3 He4 ⟶ C12 + 𝛾: 2.247437734629482e-14}
We see that we get the same value as computing it manually.
