Dealing with Duplicate Rates#
Sometimes we combine multiple sources of rates into a RateCollection or network, and we run the risk of
having the same rate sequence duplicated, even though the rate itself may be different (i.e., a different
source or tabulated vs. ReacLib).
import pynucastro as pyna
rl = pyna.ReacLibLibrary()
wl = pyna.TabularLibrary()
Let’s create a set of nuclei that can describe basic C burning
nuclei = ["c12", "o16", "ne20",
"na23", "mg23", "mg24",
"p", "n", "a"]
We’ll pull rates in both from the ReacLib library and from our tabulated weak rates
lib = rl.linking_nuclei(nuclei) + wl.linking_nuclei(nuclei)
warning: C12 was not able to be linked in TabularLibrary
warning: He4 was not able to be linked in TabularLibrary
warning: O16 was not able to be linked in TabularLibrary
warning: Ne20 was not able to be linked in TabularLibrary
warning: Mg24 was not able to be linked in TabularLibrary
If we try to create a network or RateCollection now, it will fail:
rc = pyna.RateCollection(libraries=[lib])
[[n ⟶ p + e⁻ + 𝜈, n ⟶ p + e⁻ + 𝜈], [Mg23 ⟶ Na23 + e⁺ + 𝜈, Mg23 + e⁻ ⟶ Na23 + 𝜈]]
---------------------------------------------------------------------------
RateDuplicationError Traceback (most recent call last)
Cell In[5], line 1
----> 1 rc = pyna.RateCollection(libraries=[lib])
File /opt/hostedtoolcache/Python/3.14.3/x64/lib/python3.14/site-packages/pynucastro/networks/rate_collection.py:737, in RateCollection.__init__(self, rate_files, libraries, rates, inert_nuclei, do_screening, verbose)
733 combined_library += lib
735 self.rates = self.rates + combined_library.get_rates()
--> 737 self._build_collection()
File /opt/hostedtoolcache/Python/3.14.3/x64/lib/python3.14/site-packages/pynucastro/networks/rate_collection.py:870, in RateCollection._build_collection(self)
868 if dupes := self.find_duplicate_links():
869 print(dupes)
--> 870 raise RateDuplicationError("Duplicate rates found")
RateDuplicationError: Duplicate rates found
We see that we get a RateDuplicationError because one or more rates has a duplicate.
We can find the duplicates using the find_duplicate_links method. This will return a list of lists of rates that all provide the same link.
lib.find_duplicate_links()
[[n ⟶ p + e⁻ + 𝜈, n ⟶ p + e⁻ + 𝜈],
[Mg23 ⟶ Na23 + e⁺ + 𝜈, Mg23 + e⁻ ⟶ Na23 + 𝜈]]
We see that the neutron decay and the decay of \({}^{23}\mathrm{Mg}\) are both duplicated. Let’s see if we can learn more:
for n, p in enumerate(lib.find_duplicate_links()):
print(f"dupe {n}:")
print(f" rate 1: {p[0]} ({type(p[0])})")
print(f" rate 2: {p[1]} ({type(p[1])})")
dupe 0:
rate 1: n ⟶ p + e⁻ + 𝜈 (<class 'pynucastro.rates.reaclib_rate.ReacLibRate'>)
rate 2: n ⟶ p + e⁻ + 𝜈 (<class 'pynucastro.rates.tabular_rate.TabularRate'>)
dupe 1:
rate 1: Mg23 ⟶ Na23 + e⁺ + 𝜈 (<class 'pynucastro.rates.reaclib_rate.ReacLibRate'>)
rate 2: Mg23 + e⁻ ⟶ Na23 + 𝜈 (<class 'pynucastro.rates.tabular_rate.TabularRate'>)
In this case, we see that one is a ReacLibRate and the other is a TabularRate.
The first duplicate is neutron decay. ReacLib simply has the decay constant, with no density or temperature dependence. The tabulated rate in this case comes from the Langanke and Martínez-Pinedo [2001] and is accurate at high densities / temperatures when degeneracy effects are important.
For the second rate, note that in the TabularRate we combine the rates:
into a single effective rate. This means that our TabularRate version
of the \({}^{23}\mathrm{Mg}\) decay will contain both the \(\beta+\) decay and electron capture. Furthermore, the tabular rates have both the density and temperature dependence, and thus should be preferred to the ReacLibRates when we have a duplicate.
Here we loop over the duplicates and store the rates (the ReacLib versions) that we wish to remove in a list
rates_to_remove = []
for pair in lib.find_duplicate_links():
for r in pair:
if isinstance(r, pyna.rates.ReacLibRate):
rates_to_remove.append(r)
Now we remove the duplicates from the list
for r in rates_to_remove:
lib.remove_rate(r)
Tip
Eliminating duplicates is a common process, so a Library
has a method to automate this, eliminate_duplicates. We can achieve the same result as above by doing:
lib.eliminate_duplicates()
Finally, we can make the RateCollection without error:
rc = pyna.RateCollection(libraries=[lib])
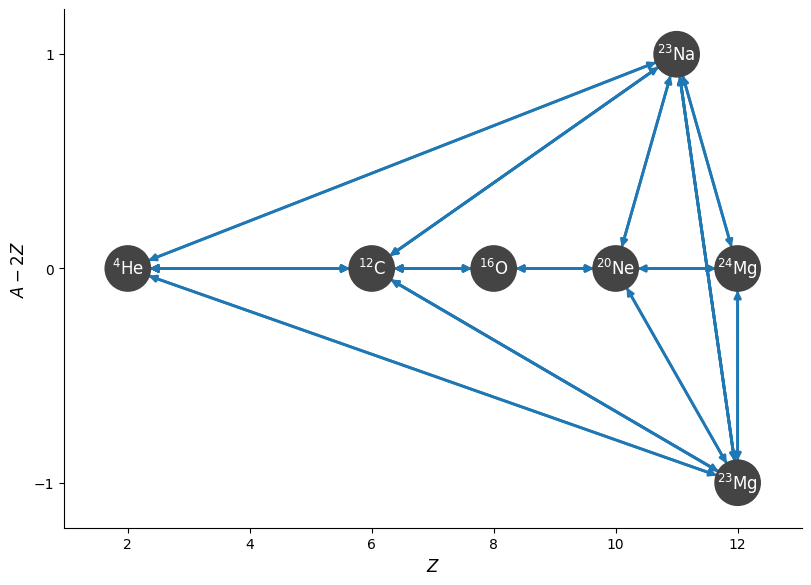
fig = rc.plot(rotated=True)