Integrating a Network#
Here we show how to create a python network and integrate it with the SciPy library.
import pynucastro as pyna
We’ll start again with the basic CNO network explored earlier. Again, we’ll read in the entire ReacLib library
and pass in the names of the rates in the form of a string A(x,y)B to filter out just the rates we are interested in.
rl = pyna.ReacLibLibrary()
rate_names = ["c12(p,g)n13",
"c13(p,g)n14",
"n13(,)c13",
"n13(p,g)o14",
"n14(p,g)o15",
"n15(p,a)c12",
"o14(,)n14",
"o15(,)n15"]
rates = rl.get_rate_by_name(rate_names)
rates
[C12 + p ⟶ N13 + 𝛾,
C13 + p ⟶ N14 + 𝛾,
N13 ⟶ C13 + e⁺ + 𝜈,
N13 + p ⟶ O14 + 𝛾,
N14 + p ⟶ O15 + 𝛾,
N15 + p ⟶ He4 + C12,
O14 ⟶ N14 + e⁺ + 𝜈,
O15 ⟶ N15 + e⁺ + 𝜈]
A PythonNetwork is based on a RateCollection but has methods to write the RHS of the system of ODEs.
pynet = pyna.PythonNetwork(rates=rates)
fig = pynet.plot()
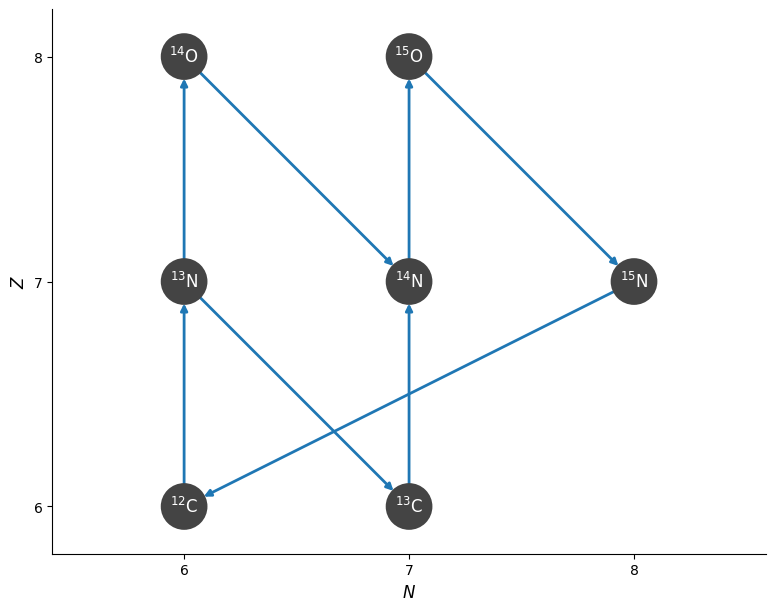
For example, this network knows how to write the full term for a reaction that goes into the \(dY/dt\) equation of the ODE system.
Here we pick one of the rates that is part of the network an explore it.
r = pynet.rates[1]
print(r)
C13 + p ⟶ N14 + 𝛾
a rate also knows what its contribution is to the \(dY/dt\) equation is (through the ydot_string_py method):
print(r.ydot_string_py())
rho*Y[jp]*Y[jc13]*rate_eval.p_C13_to_N14_reaclib
and the python code needed to evaluate this rate (the T-dependent part) is constructed by the ReacLibRate class function_string_py method:
print(r.function_string_py())
@numba.njit()
def p_C13_to_N14_reaclib(rate_eval, tf, log_scor=0.0):
# C13 + p --> N14
rate = 0.0
# nacrn
ln_set_rate = 18.5155 + -13.72*tf.T913i + -0.450018*tf.T913 \
+ 3.70823*tf.T9 + -1.70545*tf.T953 + -0.666667*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# nacrr
ln_set_rate = 13.9637 + -5.78147*tf.T9i + -0.196703*tf.T913 \
+ 0.142126*tf.T9 + -0.0238912*tf.T953 + -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# nacrr
ln_set_rate = 15.1825 + -13.5543*tf.T9i \
+ -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.p_C13_to_N14_reaclib = rate
Note
The temperature-dependent rate evaluation functions take a Tfactor object, which precomputes most of the commonly-used temperature factors in the rates.
Note
Other rate classes (like TabularRate
or ApproximateRate) have their own implementations of function_string_py().
The write_network method will output the python code needed to define the RHS of a network for integration with the SciPy integrators.
Tip
Since python code can be slow, we use Numba to do just-in-time compilation of the functions to speed things up. The numba package needs to be installed to take advantage of this.
pynet.write_network("cno_test_integrate.py")
%cat cno_test_integrate.py
import numba
import numpy as np
from pynucastro.constants import constants
from numba.experimental import jitclass
from pynucastro.rates import (TableIndex, TableInterpolator, TabularRate,
TempTableInterpolator, TemperatureTabularRate,
Tfactors)
from pynucastro.screening import PlasmaState, ScreenFactors
jp = 0
jhe4 = 1
jc12 = 2
jc13 = 3
jn13 = 4
jn14 = 5
jn15 = 6
jo14 = 7
jo15 = 8
nnuc = 9
A = np.zeros((nnuc), dtype=np.int32)
A[jp] = 1
A[jhe4] = 4
A[jc12] = 12
A[jc13] = 13
A[jn13] = 13
A[jn14] = 14
A[jn15] = 15
A[jo14] = 14
A[jo15] = 15
Z = np.zeros((nnuc), dtype=np.int32)
Z[jp] = 1
Z[jhe4] = 2
Z[jc12] = 6
Z[jc13] = 6
Z[jn13] = 7
Z[jn14] = 7
Z[jn15] = 7
Z[jo14] = 8
Z[jo15] = 8
# masses in ergs
mass = np.zeros((nnuc), dtype=np.float64)
mass[jp] = 0.0015040963047307696
mass[jhe4] = 0.0059735574859708365
mass[jc12] = 0.017909017027273523
mass[jc13] = 0.01940644192976114
mass[jn13] = 0.01940999951603316
mass[jn14] = 0.020898440897976135
mass[jn15] = 0.022386433805845516
mass[jo14] = 0.020906683078094165
mass[jo15] = 0.02239084645968795
names = []
names.append("H1")
names.append("He4")
names.append("C12")
names.append("C13")
names.append("N13")
names.append("N14")
names.append("N15")
names.append("O14")
names.append("O15")
def to_composition(Y):
"""Convert an array of molar fractions to a Composition object."""
from pynucastro import Composition, Nucleus
nuclei = [Nucleus.from_cache(name) for name in names]
comp = Composition(nuclei)
for i, nuc in enumerate(nuclei):
comp.X[nuc] = Y[i] * A[i]
return comp
def energy_release(dY):
"""return the energy release in erg/g (/s if dY is actually dY/dt)"""
enuc = 0.0
for i, y in enumerate(dY):
enuc += y * mass[i]
enuc *= -1*constants.N_A
return enuc
@jitclass([
("p_C12_to_N13_reaclib", numba.float64),
("p_C13_to_N14_reaclib", numba.float64),
("N13_to_C13_reaclib", numba.float64),
("p_N13_to_O14_reaclib", numba.float64),
("p_N14_to_O15_reaclib", numba.float64),
("p_N15_to_He4_C12_reaclib", numba.float64),
("O14_to_N14_reaclib", numba.float64),
("O15_to_N15_reaclib", numba.float64),
])
class RateEval:
def __init__(self):
self.p_C12_to_N13_reaclib = np.nan
self.p_C13_to_N14_reaclib = np.nan
self.N13_to_C13_reaclib = np.nan
self.p_N13_to_O14_reaclib = np.nan
self.p_N14_to_O15_reaclib = np.nan
self.p_N15_to_He4_C12_reaclib = np.nan
self.O14_to_N14_reaclib = np.nan
self.O15_to_N15_reaclib = np.nan
@numba.njit()
def ye(Y):
return np.sum(Z * Y)/np.sum(A * Y)
@numba.njit()
def p_C12_to_N13_reaclib(rate_eval, tf, log_scor=0.0):
# C12 + p --> N13
rate = 0.0
# ls09n
ln_set_rate = 17.1482 + -13.692*tf.T913i + -0.230881*tf.T913 \
+ 4.44362*tf.T9 + -3.15898*tf.T953 + -0.666667*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# ls09r
ln_set_rate = 17.5428 + -3.77849*tf.T9i + -5.10735*tf.T913i + -2.24111*tf.T913 \
+ 0.148883*tf.T9 + -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.p_C12_to_N13_reaclib = rate
@numba.njit()
def p_C13_to_N14_reaclib(rate_eval, tf, log_scor=0.0):
# C13 + p --> N14
rate = 0.0
# nacrn
ln_set_rate = 18.5155 + -13.72*tf.T913i + -0.450018*tf.T913 \
+ 3.70823*tf.T9 + -1.70545*tf.T953 + -0.666667*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# nacrr
ln_set_rate = 13.9637 + -5.78147*tf.T9i + -0.196703*tf.T913 \
+ 0.142126*tf.T9 + -0.0238912*tf.T953 + -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# nacrr
ln_set_rate = 15.1825 + -13.5543*tf.T9i \
+ -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.p_C13_to_N14_reaclib = rate
@numba.njit()
def N13_to_C13_reaclib(rate_eval, tf, log_scor=0.0):
# N13 --> C13
rate = 0.0
# wc12w
ln_set_rate = -6.7601
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.N13_to_C13_reaclib = rate
@numba.njit()
def p_N13_to_O14_reaclib(rate_eval, tf, log_scor=0.0):
# N13 + p --> O14
rate = 0.0
# lg06r
ln_set_rate = 10.9971 + -6.12602*tf.T9i + 1.57122*tf.T913i \
+ -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# lg06n
ln_set_rate = 18.1356 + -15.1676*tf.T913i + 0.0955166*tf.T913 \
+ 3.0659*tf.T9 + -0.507339*tf.T953 + -0.666667*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.p_N13_to_O14_reaclib = rate
@numba.njit()
def p_N14_to_O15_reaclib(rate_eval, tf, log_scor=0.0):
# N14 + p --> O15
rate = 0.0
# im05r
ln_set_rate = 6.73578 + -4.891*tf.T9i \
+ 0.0682*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# im05r
ln_set_rate = 7.65444 + -2.998*tf.T9i \
+ -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# im05n
ln_set_rate = 20.1169 + -15.193*tf.T913i + -4.63975*tf.T913 \
+ 9.73458*tf.T9 + -9.55051*tf.T953 + 0.333333*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# im05n
ln_set_rate = 17.01 + -15.193*tf.T913i + -0.161954*tf.T913 \
+ -7.52123*tf.T9 + -0.987565*tf.T953 + -0.666667*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.p_N14_to_O15_reaclib = rate
@numba.njit()
def p_N15_to_He4_C12_reaclib(rate_eval, tf, log_scor=0.0):
# N15 + p --> He4 + C12
rate = 0.0
# nacrn
ln_set_rate = 27.4764 + -15.253*tf.T913i + 1.59318*tf.T913 \
+ 2.4479*tf.T9 + -2.19708*tf.T953 + -0.666667*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# nacrr
ln_set_rate = -6.57522 + -1.1638*tf.T9i + 22.7105*tf.T913 \
+ -2.90707*tf.T9 + 0.205754*tf.T953 + -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# nacrr
ln_set_rate = 20.8972 + -7.406*tf.T9i \
+ -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
# nacrr
ln_set_rate = -4.87347 + -2.02117*tf.T9i + 30.8497*tf.T913 \
+ -8.50433*tf.T9 + -1.54426*tf.T953 + -1.5*tf.lnT9
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.p_N15_to_He4_C12_reaclib = rate
@numba.njit()
def O14_to_N14_reaclib(rate_eval, tf, log_scor=0.0):
# O14 --> N14
rate = 0.0
# wc12w
ln_set_rate = -4.62354
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.O14_to_N14_reaclib = rate
@numba.njit()
def O15_to_N15_reaclib(rate_eval, tf, log_scor=0.0):
# O15 --> N15
rate = 0.0
# wc12w
ln_set_rate = -5.17053
ln_set_rate += log_scor
set_rate = np.exp(ln_set_rate)
rate += set_rate
rate_eval.O15_to_N15_reaclib = rate
def rhs(t, Y, rho, T, screen_func=None):
return rhs_eq(t, Y, rho, T, screen_func)
@numba.njit()
def rhs_eq(t, Y, rho, T, screen_func):
tf = Tfactors(T)
rate_eval = RateEval()
log_scor_p_C12 = 0.0
log_scor_p_C13 = 0.0
log_scor_p_N13 = 0.0
log_scor_p_N14 = 0.0
log_scor_p_N15 = 0.0
if screen_func is not None:
plasma_state = PlasmaState(T, rho, Y, Z)
scn_fac = ScreenFactors(1, 1, 6, 12)
log_scor_p_C12 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 6, 13)
log_scor_p_C13 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 7, 13)
log_scor_p_N13 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 7, 14)
log_scor_p_N14 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 7, 15)
log_scor_p_N15 = screen_func(plasma_state, scn_fac)
# reaclib rates
p_C12_to_N13_reaclib(rate_eval, tf, log_scor=log_scor_p_C12)
p_C13_to_N14_reaclib(rate_eval, tf, log_scor=log_scor_p_C13)
N13_to_C13_reaclib(rate_eval, tf)
p_N13_to_O14_reaclib(rate_eval, tf, log_scor=log_scor_p_N13)
p_N14_to_O15_reaclib(rate_eval, tf, log_scor=log_scor_p_N14)
p_N15_to_He4_C12_reaclib(rate_eval, tf, log_scor=log_scor_p_N15)
O14_to_N14_reaclib(rate_eval, tf)
O15_to_N15_reaclib(rate_eval, tf)
dYdt = np.zeros((nnuc), dtype=np.float64)
dYdt[jp] = (
-rho*Y[jp]*Y[jc12]*rate_eval.p_C12_to_N13_reaclib +
-rho*Y[jp]*Y[jc13]*rate_eval.p_C13_to_N14_reaclib +
-rho*Y[jp]*Y[jn13]*rate_eval.p_N13_to_O14_reaclib +
-rho*Y[jp]*Y[jn14]*rate_eval.p_N14_to_O15_reaclib +
-rho*Y[jp]*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib
)
dYdt[jhe4] = (
+rho*Y[jp]*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib
)
dYdt[jc12] = (
-rho*Y[jp]*Y[jc12]*rate_eval.p_C12_to_N13_reaclib +
+rho*Y[jp]*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib
)
dYdt[jc13] = (
-rho*Y[jp]*Y[jc13]*rate_eval.p_C13_to_N14_reaclib +
+Y[jn13]*rate_eval.N13_to_C13_reaclib
)
dYdt[jn13] = (
+rho*Y[jp]*Y[jc12]*rate_eval.p_C12_to_N13_reaclib +
-Y[jn13]*rate_eval.N13_to_C13_reaclib +
-rho*Y[jp]*Y[jn13]*rate_eval.p_N13_to_O14_reaclib
)
dYdt[jn14] = (
+rho*Y[jp]*Y[jc13]*rate_eval.p_C13_to_N14_reaclib +
-rho*Y[jp]*Y[jn14]*rate_eval.p_N14_to_O15_reaclib +
+Y[jo14]*rate_eval.O14_to_N14_reaclib
)
dYdt[jn15] = (
-rho*Y[jp]*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib +
+Y[jo15]*rate_eval.O15_to_N15_reaclib
)
dYdt[jo14] = (
+rho*Y[jp]*Y[jn13]*rate_eval.p_N13_to_O14_reaclib +
-Y[jo14]*rate_eval.O14_to_N14_reaclib
)
dYdt[jo15] = (
+rho*Y[jp]*Y[jn14]*rate_eval.p_N14_to_O15_reaclib +
-Y[jo15]*rate_eval.O15_to_N15_reaclib
)
return dYdt
def jacobian(t, Y, rho, T, screen_func=None):
return jacobian_eq(t, Y, rho, T, screen_func)
@numba.njit()
def jacobian_eq(t, Y, rho, T, screen_func):
tf = Tfactors(T)
rate_eval = RateEval()
log_scor_p_C12 = 0.0
log_scor_p_C13 = 0.0
log_scor_p_N13 = 0.0
log_scor_p_N14 = 0.0
log_scor_p_N15 = 0.0
if screen_func is not None:
plasma_state = PlasmaState(T, rho, Y, Z)
scn_fac = ScreenFactors(1, 1, 6, 12)
log_scor_p_C12 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 6, 13)
log_scor_p_C13 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 7, 13)
log_scor_p_N13 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 7, 14)
log_scor_p_N14 = screen_func(plasma_state, scn_fac)
scn_fac = ScreenFactors(1, 1, 7, 15)
log_scor_p_N15 = screen_func(plasma_state, scn_fac)
# reaclib rates
p_C12_to_N13_reaclib(rate_eval, tf, log_scor=log_scor_p_C12)
p_C13_to_N14_reaclib(rate_eval, tf, log_scor=log_scor_p_C13)
N13_to_C13_reaclib(rate_eval, tf)
p_N13_to_O14_reaclib(rate_eval, tf, log_scor=log_scor_p_N13)
p_N14_to_O15_reaclib(rate_eval, tf, log_scor=log_scor_p_N14)
p_N15_to_He4_C12_reaclib(rate_eval, tf, log_scor=log_scor_p_N15)
O14_to_N14_reaclib(rate_eval, tf)
O15_to_N15_reaclib(rate_eval, tf)
jac = np.zeros((nnuc, nnuc), dtype=np.float64)
jac[jp, jp] = (
-rho*Y[jc12]*rate_eval.p_C12_to_N13_reaclib
-rho*Y[jc13]*rate_eval.p_C13_to_N14_reaclib
-rho*Y[jn13]*rate_eval.p_N13_to_O14_reaclib
-rho*Y[jn14]*rate_eval.p_N14_to_O15_reaclib
-rho*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jp, jc12] = (
-rho*Y[jp]*rate_eval.p_C12_to_N13_reaclib
)
jac[jp, jc13] = (
-rho*Y[jp]*rate_eval.p_C13_to_N14_reaclib
)
jac[jp, jn13] = (
-rho*Y[jp]*rate_eval.p_N13_to_O14_reaclib
)
jac[jp, jn14] = (
-rho*Y[jp]*rate_eval.p_N14_to_O15_reaclib
)
jac[jp, jn15] = (
-rho*Y[jp]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jhe4, jp] = (
+rho*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jhe4, jn15] = (
+rho*Y[jp]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jc12, jp] = (
-rho*Y[jc12]*rate_eval.p_C12_to_N13_reaclib
+rho*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jc12, jc12] = (
-rho*Y[jp]*rate_eval.p_C12_to_N13_reaclib
)
jac[jc12, jn15] = (
+rho*Y[jp]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jc13, jp] = (
-rho*Y[jc13]*rate_eval.p_C13_to_N14_reaclib
)
jac[jc13, jc13] = (
-rho*Y[jp]*rate_eval.p_C13_to_N14_reaclib
)
jac[jc13, jn13] = (
+rate_eval.N13_to_C13_reaclib
)
jac[jn13, jp] = (
-rho*Y[jn13]*rate_eval.p_N13_to_O14_reaclib
+rho*Y[jc12]*rate_eval.p_C12_to_N13_reaclib
)
jac[jn13, jc12] = (
+rho*Y[jp]*rate_eval.p_C12_to_N13_reaclib
)
jac[jn13, jn13] = (
-rate_eval.N13_to_C13_reaclib
-rho*Y[jp]*rate_eval.p_N13_to_O14_reaclib
)
jac[jn14, jp] = (
-rho*Y[jn14]*rate_eval.p_N14_to_O15_reaclib
+rho*Y[jc13]*rate_eval.p_C13_to_N14_reaclib
)
jac[jn14, jc13] = (
+rho*Y[jp]*rate_eval.p_C13_to_N14_reaclib
)
jac[jn14, jn14] = (
-rho*Y[jp]*rate_eval.p_N14_to_O15_reaclib
)
jac[jn14, jo14] = (
+rate_eval.O14_to_N14_reaclib
)
jac[jn15, jp] = (
-rho*Y[jn15]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jn15, jn15] = (
-rho*Y[jp]*rate_eval.p_N15_to_He4_C12_reaclib
)
jac[jn15, jo15] = (
+rate_eval.O15_to_N15_reaclib
)
jac[jo14, jp] = (
+rho*Y[jn13]*rate_eval.p_N13_to_O14_reaclib
)
jac[jo14, jn13] = (
+rho*Y[jp]*rate_eval.p_N13_to_O14_reaclib
)
jac[jo14, jo14] = (
-rate_eval.O14_to_N14_reaclib
)
jac[jo15, jp] = (
+rho*Y[jn14]*rate_eval.p_N14_to_O15_reaclib
)
jac[jo15, jn14] = (
+rho*Y[jp]*rate_eval.p_N14_to_O15_reaclib
)
jac[jo15, jo15] = (
-rate_eval.O15_to_N15_reaclib
)
return jac
We can now import the network that was just created and integrate it using the SciPy ODE solvers
import cno_test_integrate as cno
Integrating the network#
We can use the stiff ODE integration solvers that are part of SciPy solve_ivp to integrate this system.
from scipy.integrate import solve_ivp
import numpy as np
Initialize the thermodynamic conditions and initial composition. We express the composition as molar fractions, Y0.
rho = 150
T = 1.5e7
X0 = np.zeros(cno.nnuc)
X0[cno.jp] = 0.7
X0[cno.jhe4] = 0.28
X0[cno.jc12] = 0.02
Y0 = X0/cno.A
Now we integrate. For our stiff networks, the BDF method works well.
tmax = 1.e20
sol = solve_ivp(cno.rhs, [0, tmax], Y0, method="BDF", jac=cno.jacobian,
dense_output=True, args=(rho, T), rtol=1.e-6, atol=1.e-6)
Tip
You may need to change the relative (rtol) or absolute (atol) tolerances if you want to more accurately capture trace nuclei. Making them smaller will make the integrator work harder.
Plotting the results#
import matplotlib.pyplot as plt
fig = plt.figure()
ax = fig.add_subplot(111)
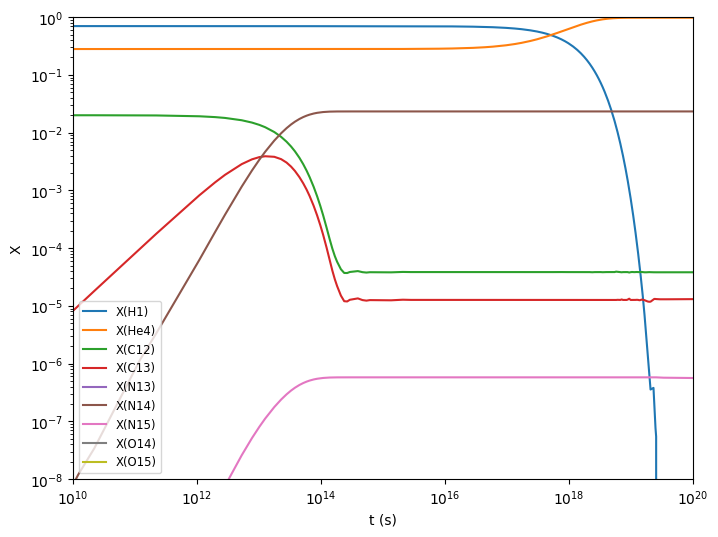
for i in range(cno.nnuc):
ax.loglog(sol.t, sol.y[i,:] * cno.A[i], label=f"X({cno.names[i].capitalize()})")
ax.set_xlim(1.e10, 1.e20)
ax.set_ylim(1.e-8, 1.0)
ax.legend(fontsize="small")
ax.set_xlabel("t (s)")
ax.set_ylabel("X")
fig.set_size_inches((8, 6))
Energy release#
We can get the energy release from the change in molar abundances, \(\Delta Y\). This will be computed as:
Here \(M_i\) is the mass of the nucleus \(i\) (this is tabulated in
the network as mass[]).
E = cno.energy_release(sol.y[:,-1] - Y0)
print(f"E = {E:20.10g}")
E = 4.510855593e+18
This result is in erg/g
We can compute the instantaneous energy generation rate as well,
epsilon = cno.energy_release(cno.rhs(0.0, Y0, rho, T))
print(f"epsilon = {epsilon:20.10g}")
epsilon = 117.7738358
