Binding Energy per Nucleon#
We can explore and plot the binding energy per nucleon to understand when fusion and fission operate.
import pynucastro as pyna
First we’ll get all nuclei with known masses and look at the binding energy
nuclei = pyna.get_all_nuclei()
len(nuclei)
3558
We see there are > 3500 nuclei with measured masses
Most tightly bound nucleus#
We can easily find the nucleus that is most tightly bound
nuc_bound = max(nuclei, key=lambda n : n.nucbind)
nuc_bound
Ni62
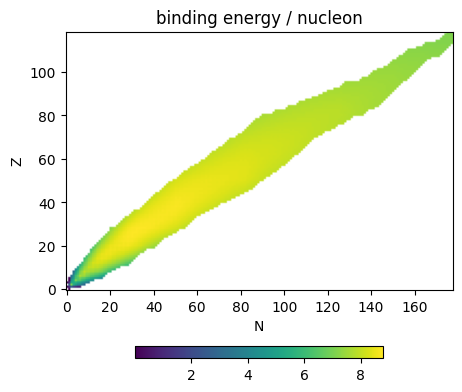
Binding energy plot#
Now we can make a plot of binding energy per nucleon for all nuclei
As = [n.A for n in nuclei]
BEs = [n.nucbind for n in nuclei]
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.scatter(As, BEs, s=5)
ax.set_xlabel("number of nucleons")
ax.set_ylabel("binding energy per nucleon (MeV)")
Text(0, 0.5, 'binding energy per nucleon (MeV)')
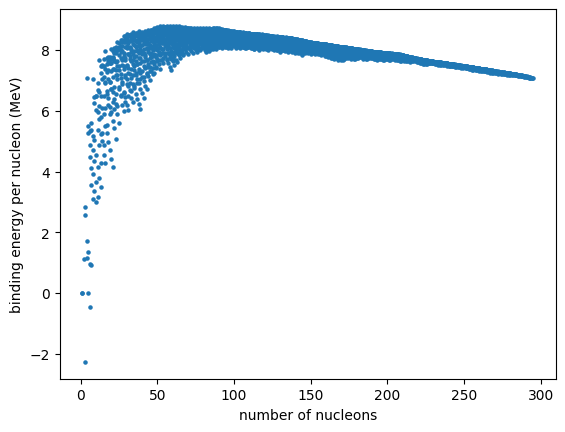
Cleaner plot#
We see that there is quite a spread in binding energy for each nucleon count. We can instead consider only the most tightly bound nucleus at each mass number. We will also only consider those that are stable (or have half lives > 1 million years)
max_A = max(As)
max_A
295
nuc = []
1 million years in seconds
million_years = 1.e6 * 365.25 * 24 * 3600
for A in range(1, max_A+1):
# for mass number A, find the nucleus with the maximum binding energy
try:
_new = max((n for n in nuclei
if n.A == A and (n.tau == "stable" or
(n.tau is not None and n.tau > million_years))),
key=lambda q: q.nucbind)
except ValueError:
# no stable nucleus of this mass
continue
nuc.append(_new)
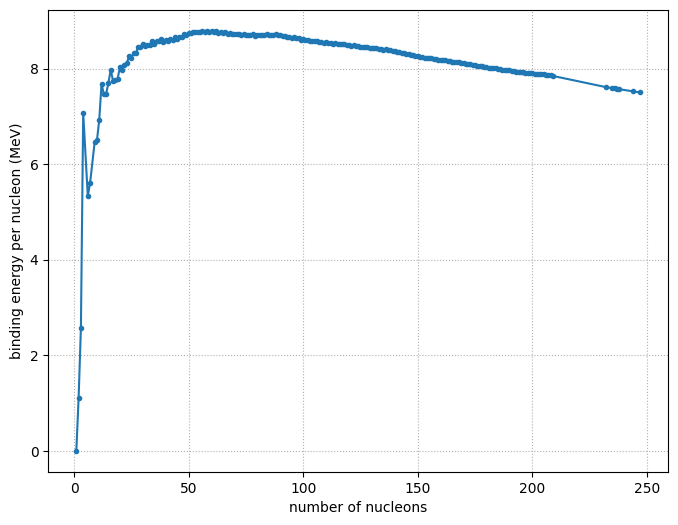
We now plot just these most tightly bound nuclei
As = [n.A for n in nuc]
BEs = [n.nucbind for n in nuc]
fig, ax = plt.subplots()
ax.plot(As, BEs, marker="o", markersize="3")
ax.set_xlabel("number of nucleons")
ax.set_ylabel("binding energy per nucleon (MeV)")
ax.grid(ls=":")
fig.set_size_inches((8, 6))
Visualizing as function of (N, Z)#
We want to visualize the mass excess and binding energy in the \(Z\)-\(N\) plane. First let’s get the extent of \(N\) and \(Z\) in our nucleus list.
max_Z = max(nuclei, key=lambda n : n.Z).Z
max_N = max(nuclei, key=lambda n : n.N).N
and the maximum absolute value of the mass excess (in MeV)
dm_mag = abs(max(nuclei, key=lambda n: abs(n.dm)).dm)
dm_mag
201.37
Now we’ll create an array to store dm(Z, N) and be(Z, N) and loop over all the nuclei and store each mass excess and binding energy / nucleon.
import numpy as np
dm = np.zeros((max_Z+1, max_N+1))
be = np.zeros((max_Z+1, max_N+1))
We’ll initialize these to NaN so we can mask out the regions where there are no nuclei
dm[:,:] = np.nan
be[:,:] = np.nan
for n in nuclei:
dm[n.Z, n.N] = n.dm
be[n.Z, n.N] = n.nucbind
Note
Due to mass excess estimation in the nuclear databases, a few nuclei have negative binding energies.
n = [n for n in nuclei if n.nucbind < 0]
n
[Li3, B6]
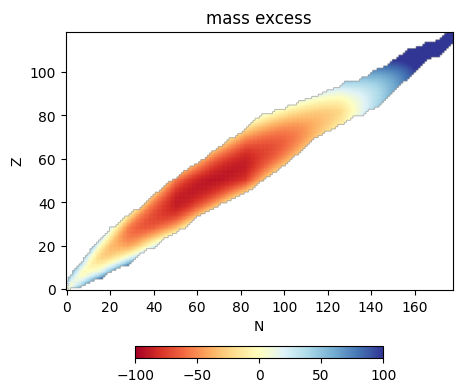
Finally, we can plot
import matplotlib as mpl
# mask out the regions with no nuclei
cmap = mpl.colormaps['RdYlBu']
cmap.set_bad(color='white')
fig, ax = plt.subplots()
im = ax.imshow(dm, origin="lower", cmap="RdYlBu",
vmin=-100, vmax=100)
ax.set_xlabel("N")
ax.set_ylabel("Z")
ax.set_title("mass excess")
fig.colorbar(im, ax=ax, location="bottom", shrink=0.5)
<matplotlib.colorbar.Colorbar at 0x7f955b6a78c0>
# mask out the regions with no nuclei
cmap = mpl.colormaps['viridis']
cmap.set_bad(color='white')
fig, ax = plt.subplots()
im = ax.imshow(be, origin="lower", cmap=cmap,
vmin=0.01, vmax=np.nanmax(be))
ax.set_xlabel("N")
ax.set_ylabel("Z")
ax.set_title("binding energy / nucleon")
fig.colorbar(im, ax=ax, location="bottom", shrink=0.5)
<matplotlib.colorbar.Colorbar at 0x7f955b57b4d0>