Screening Rates#
Consider nuclear fusion between two ions surrounded by a sea of electrons. The electron screening effect reduces the effective Coulomb potential barrier that the ions must overcome for fusion to take place, causing an enhancement to the nuclear reaction rates. Suppose the unscreened rate is represented by \(R_\mathrm{uns}\), then screened rate is
where \(h\) characterizes the screening magnitude. pynucastro has various screening implementations that computes screening magnitude, \(h\).
Screening implementations#
pynucastro currently has 4 screening implementations:
potekhin_1998based on Chabrier and Potekhin [1998]chugunov_2007based on Chugunov et al. [2007]chugunov_2009based on Chugunov and DeWitt [2009]
Each of these use various approximations to account for the influence of the plasma on screening the electric charge of the nuclei that are fusing.
Using screening#
To compute the screening factor for a rate, we need to know the entire composition (since the electron number density matters) as well as the two nuclei involved in the reaction. There are two special types that store this information:
PlasmaState: stores the aggregate information about the entire plasmaScreenFactors: stores the information about the two nuclei
We create these objects separate before we call the screening routine since there are some computational expensive calculations here that don’t change with repeated calls.
Here we create a PlasmaState that knows about the conditions where the screening is taking place and then we use that to compute the screening factor for a rate. The helper function
make_plasma_state can create the PlasmaState object
import math
import pynucastro as pyna
nuclei = [pyna.Nucleus("h1"),
pyna.Nucleus("he4"),
pyna.Nucleus("c12"),
pyna.Nucleus("o16"),
pyna.Nucleus("n14"),
pyna.Nucleus("ca40")]
comp = pyna.Composition(nuclei)
comp.set_solar_like()
dens = 1.e6
temp = 1.e8
plasma = pyna.make_plasma_state(temp, dens, comp.get_molar())
Now let’s get the \({}^{12}\mathrm{C}(\alpha,\gamma){}^{16}\mathrm{O}\) rate and compute the screening factor
reaclib_library = pyna.ReacLibLibrary()
rfilter = pyna.RateFilter(reactants=["c12", "he4"], products=["o16"])
r = reaclib_library.filter(rfilter).get_rates()[0]
r
C12 + He4 ⟶ O16 + 𝛾
For the rate, we need the ScreenFactors object
scn_fac = pyna.make_screen_factors(r.ion_screen[0], r.ion_screen[1])
Finally, we’ll select the Chugunov (2009) screening and compute the screening factor. All of the needed thermodynamic information is contained in the PlasmaState and all of the needed
reaction rate information is contained in the ScreenFactors
Note
All screening routines return \(h\) = log(screening). Therefore, in order to get the actual multiplicative rate enhancement factor, we need to do an exp() afterwards.
from pynucastro.screening import chugunov_2009
log_scn = chugunov_2009(plasma, scn_fac)
scn = math.exp(log_scn)
print(f"{scn:12.8f}")
4.42076841
In this case, we see that screening enhances the rate by over 4 times!
Screening map#
For a RateCollection or a network derived from it, there are a lot of rates that will need to be screened, and some might have the same nuclei that need to be screened. A “screening map” keeps track of all of the rates that need to be screened for the same set of reactants.
The screening map is a list of ScreeningPair objects which contain the pair of nuclei that need a screening factor computed as well as the list of all
the rates that screening factor applies to.
Here’s an example: let’s build a helium and carbon burning network.
mynet = reaclib_library.linking_nuclei(["p", "n", "he4", "c12", "o16",
"na23", "mg24", "ne20"])
pynet = pyna.PythonNetwork(libraries=[mynet])
From the RateCollection, we can get the screening map using the get_screening_map function:
from pynucastro.screening import get_screening_map
screen_map = get_screening_map(pynet.get_rates())
for s in screen_map:
print(s)
screening for He4 + C12
rates:
C12 + He4 ⟶ O16 + 𝛾
screening for He4 + O16
rates:
O16 + He4 ⟶ Ne20 + 𝛾
screening for He4 + Ne20
rates:
Ne20 + He4 ⟶ Mg24 + 𝛾
Ne20 + He4 ⟶ p + Na23
Ne20 + He4 ⟶ C12 + C12
screening for p + Na23
rates:
Na23 + p ⟶ Mg24 + 𝛾
Na23 + p ⟶ He4 + Ne20
Na23 + p ⟶ C12 + C12
screening for C12 + C12
rates:
C12 + C12 ⟶ p + Na23
C12 + C12 ⟶ He4 + Ne20
screening for C12 + O16
rates:
O16 + C12 ⟶ He4 + Mg24
screening for He4 + Mg24
rates:
Mg24 + He4 ⟶ C12 + O16
screening for He4 + He4
rates:
3 He4 ⟶ C12 + 𝛾
screening for He4 + Be8
rates:
3 He4 ⟶ C12 + 𝛾
Here we see that the screening for some pairs of nuclei (like \(p + {}^{23}\mathrm{Na}\)) apply to several rates.
Screening and a python network#
When we write out the module that defines a python network, it contains all of the information needed to define the righthand side and Jacobian. By default, screening is not used, but the rhs and jacobian functions can take an optional argument that is the name of the screening function to use.
Here we demonstrate this.
import numpy as np
import matplotlib.pyplot as plt
from scipy.integrate import solve_ivp
pynet.write_network("cburn.py")
import cburn
First we’ll run with screening. Here we pass the name of the screening function to the rhs and jacobian function through the args keyword
We’ll pick conditions appropriate for the center of a Chandra mass white dwarf just after a carbon flame ignites.
rho = 1.e9
T = 2.e9
X0 = np.zeros(cburn.nnuc)
X0[cburn.jc12] = 0.5
X0[cburn.jo16] = 0.5
Y0 = X0/cburn.A
tmax = 1000.0
sol = solve_ivp(cburn.rhs, [0, tmax], Y0, method="BDF", jac=cburn.jacobian,
dense_output=True, args=(rho, T, chugunov_2009), rtol=1.e-6, atol=1.e-10)
Now we run without screening:
sol_noscreen = solve_ivp(cburn.rhs, [0, tmax], Y0, method="BDF", jac=cburn.jacobian,
dense_output=True, args=(rho, T), rtol=1.e-6, atol=1.e-10)
and we can plot the two cases together. The non-screened X’s will be shown with a dotted line.
fig = plt.figure()
ax = fig.add_subplot(111)
threshold = 1.e-4
icolor = 0
for i in range(cburn.nnuc):
if (sol.y[i,:]).max() > threshold:
ax.loglog(sol.t, sol.y[i,:] * cburn.A[i],
label=f"X({cburn.names[i].capitalize()})",
color=f"C{icolor}")
ax.loglog(sol_noscreen.t, sol_noscreen.y[i,:] * cburn.A[i],
linestyle=":", color=f"C{icolor}")
icolor += 1
ax.set_ylim(1.e-8, 1.0)
ax.legend(fontsize="small")
ax.set_xlabel("t (s)")
ax.set_ylabel("X")
fig.set_size_inches((8, 6))
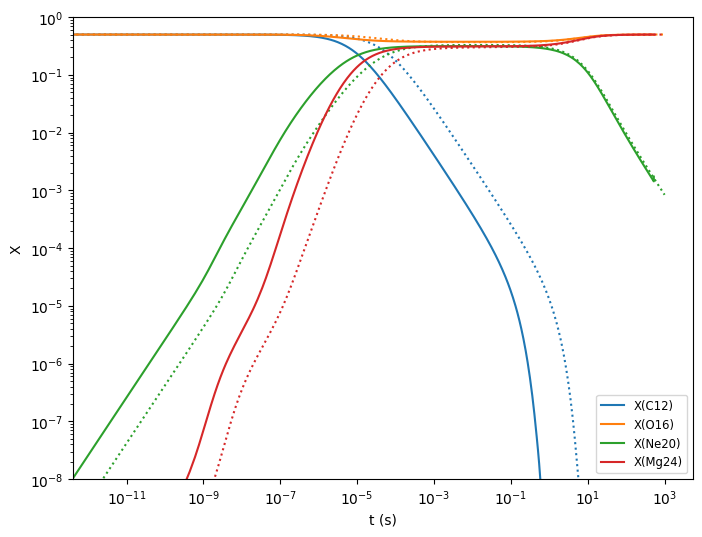
As expected, using screening makes the carbon burn much more quickly.
