Basic Networks#
The general procedure for creating a network is:
Create one or more
Libraryobjects containing the rates that you want. This can start with a list of nuclei or a list of rate names (like"c12(a,g)o16").There are multiple sources of rates in pynucastro, so sometimes several libraries will be created, filtering the desired rates from the different sources.
Remove any duplicate rates between the libraries. Usually this means keeping tabulated rates over ReacLib rates, since the tabulated rates are more general.
Rederive the inverse rates from ReacLib using detailed balance.
Create a network using either
RateCollectionor one of the classes derived from it.Approximate some of the nuclei out of the network by introducing \((\alpha, p)(p,\gamma)\) or other rate approximations.
pynucastro gives you control over each of these steps, which is needed for some complex network types. But if you just want to create a network that has all possible links connecting a set of nuclei, then the
network_helper function is enough.
Here’s an example of a CNO network with some breakout.
import pynucastro as pyna
nuclei = ["p", "he4",
"c12", "c13",
"n13", "n14", "n15",
"o14", "o15", "o16", "o17", "o18",
"f17", "f18", "f19",
"ne18", "ne19", "ne20", "ne21"]
net = pyna.network_helper(nuclei)
warning: C12 was not able to be linked in TabularLibrary
warning: O15 was not able to be linked in TabularLibrary
warning: Ne21 was not able to be linked in TabularLibrary
warning: N13 was not able to be linked in TabularLibrary
warning: He4 was not able to be linked in TabularLibrary
warning: O14 was not able to be linked in TabularLibrary
warning: Ne20 was not able to be linked in TabularLibrary
warning: C13 was not able to be linked in TabularLibrary
warning: N15 was not able to be linked in TabularLibrary
warning: p was not able to be linked in TabularLibrary
warning: O16 was not able to be linked in TabularLibrary
warning: N14 was not able to be linked in TabularLibrary
Tip
By default, network_helper will create a PythonNetwork, but
this can be changed via the network_type argument.
type(net)
pynucastro.networks.python_network.PythonNetwork
By default, network_helper will look at both ReacLibLibrary and TabularLibrary to find rates. The warnings we see above about not being able to link nuclei are from the TabularLibrary, since those nuclei do not have tabulated weak rates.
fig = net.plot(rotated=True, hide_xalpha=True)
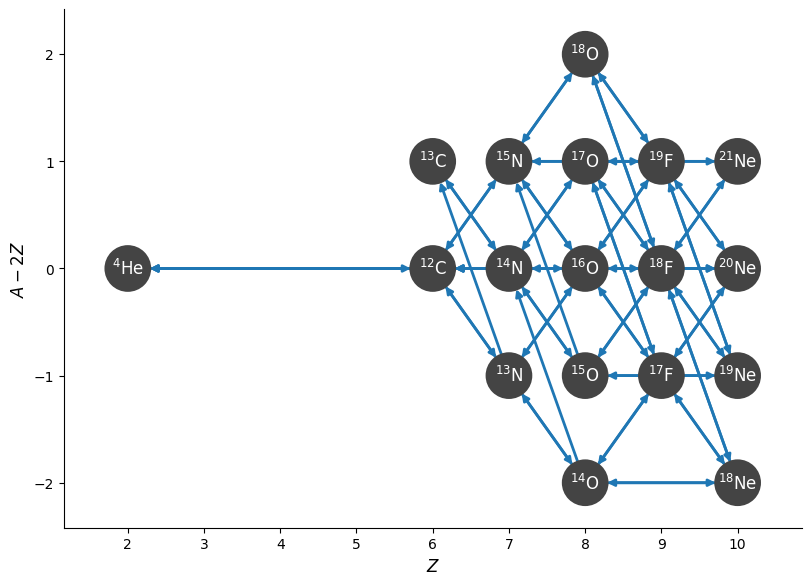
We can see that it is using some tabulated weak rates for some links:
net.summary()
Network summary
---------------
explicitly carried nuclei: 19
approximated-out nuclei: 0
inert nuclei (included in carried): 0
total number of rates: 69
rates explicitly connecting nuclei: 69
hidden rates: 0
reaclib rates: 32
weak tabular rates: 8
temperature tabular rates: 0
approximate rates: 0
derived rates: 29
modified rates: 0
custom rates: 0
We can highlight these weak rates in the plot
fig = net.plot(rotated=True, hide_xalpha=True,
highlight_filter_function=lambda r: isinstance(r, pyna.rates.TabularRate))
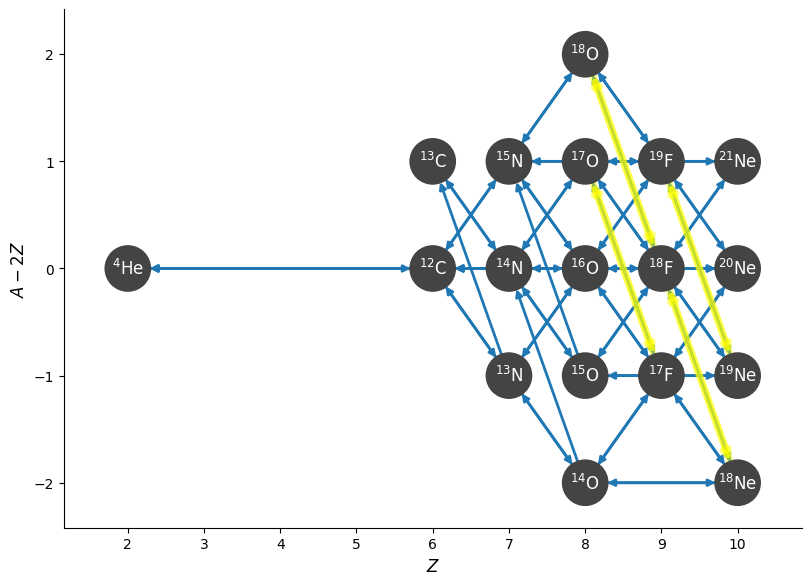
Also notice that network_helper includes both the forward and reverse rates for all links.
The notebooks following show how to have finer control when creating a network, allowing you to manually filter rates from libraries, remove duplicates, compute reverse rates, and introduce approximations.
