Modified Rates#
A ModifiedRate acts as a wrapper, encapsulating a single rate, allowing you to change the products of the stoichiometric coefficients. A good example of this comes from the aprox19 and aprox21 networks. There, an approximation to H-burning (via pp and CNO) is included, and the nucleus \({}^{14}\mathrm{N}\) is present, which is the bottleneck for CNO. Beyond that is an \(\alpha\)-chain. But \(\alpha\)-captures cannot connect \({}^{14}\mathrm{N}\) (odd \(Z\)) to the even-\(Z\) nuclei in the \(\alpha\)-chain.
To make this connection, a rate that does:
is added. For a single \({}^{14}\mathrm{N}\) we can write this as \({}^{14}\mathrm{N}(1.5\alpha,\gamma){}^{20}\mathrm{Ne}\). This is assumed to proceed at the rate for \({}^{14}\mathrm{N}(\alpha,\gamma){}^{18}\mathrm{F}\), and \({}^{18}\mathrm{F}\) is not included explicitly in the network. But we can add this approximation using
ModifiedRate.
import pynucastro as pyna
We’ll start with a basic network for He burning up to \({}^{20}\mathrm{Ne}\):
rl = pyna.ReacLibLibrary()
lib = rl.linking_nuclei(["he4", "c12", "o16", "ne20"],
with_reverse=False)
Now let’s get the \({}^{14}\mathrm{N}(1.5\alpha,\gamma){}^{20}\mathrm{Ne}\) rate and modify it to go to \({}^{20}\mathrm{Ne}\). To do this, we need to change the stoichiometric coefficients as well as the products:
n14rate = rl.get_rate_by_name("n14(a,g)f18")
new_n14 = pyna.ModifiedRate(n14rate,
new_products=[pyna.Nucleus("ne20")],
stoichiometry={pyna.Nucleus("he4"): 1.5})
new_n14
N14 + 1.5 He4 ⟶ Ne20 + 𝛾
The modified rate still knows the underlying rate we will be evaluating:
new_n14.original_rate
N14 + He4 ⟶ F18 + 𝛾
and we can see that the function to compute the rate simply evaluates the original rate and uses it.
print(new_n14.function_string_py())
@numba.njit()
def He4_N14_to_Ne20_modified(rate_eval, tf, log_scor=0.0):
# N14 + 1.5 He4 --> Ne20
He4_N14_to_F18_reaclib(rate_eval, tf, log_scor=log_scor)
rate_eval.He4_N14_to_Ne20_modified = rate_eval.He4_N14_to_F18_reaclib
Now we can add this new rate to our library:
lib.add_rate(new_n14)
and create the network from this library:
net = pyna.PythonNetwork(libraries=[lib])
The evolution for \(dY({}^{4}\mathrm{He})/dt\) also uses the correct coefficient:
print(net.full_ydot_string(pyna.Nucleus("he4")))
dYdt[jhe4] = (
-rho*Y[jhe4]*Y[jc12]*rate_eval.He4_C12_to_O16_reaclib +
-rho*Y[jhe4]*Y[jo16]*rate_eval.He4_O16_to_Ne20_reaclib +
+5.00000000000000e-01*rho*Y[jc12]**2*rate_eval.C12_C12_to_He4_Ne20_reaclib +
+ -3*1.66666666666667e-01*rho**2*Y[jhe4]**3*rate_eval.He4_He4_He4_to_C12_reaclib +
+ -1.5*rho*Y[jhe4]*Y[jn14]*rate_eval.He4_N14_to_Ne20_modified
)
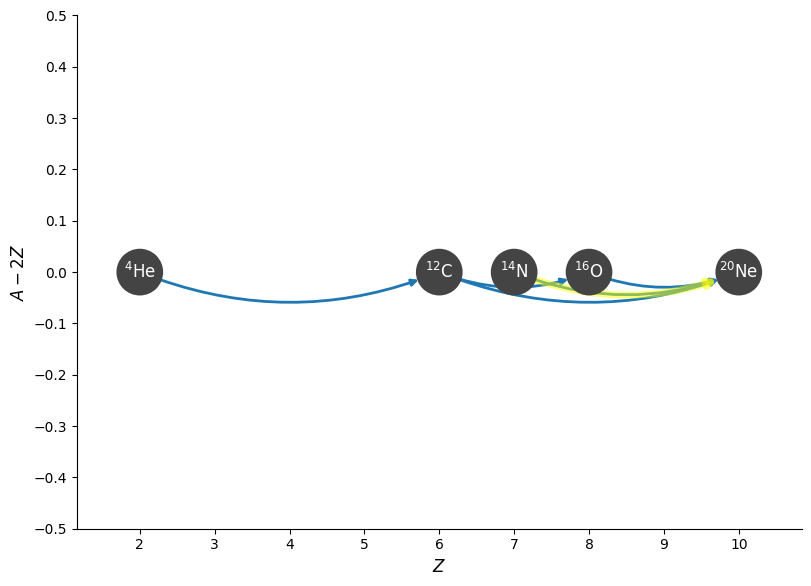
Here’s a visualization with the links from the new rate highlighted:
fig = net.plot(curved_edges=True, rotated=True, hide_xalpha=True,
highlight_filter_function=lambda r: pyna.Nucleus("n14") in r.reactants)
and the evaluation of the rates at a particular thermodynamic state:
rho = 1.e6
T = 5.e8
comp = pyna.Composition(net.unique_nuclei)
comp.set_equal()
net.evaluate_rates(rho=rho, T=T, composition=comp)
{C12 + He4 ⟶ O16 + 𝛾: 3.1635370410117465e-06,
O16 + He4 ⟶ Ne20 + 𝛾: 0.00010189935467071995,
C12 + C12 ⟶ He4 + Ne20: 2.1912364485182727e-18,
3 He4 ⟶ C12 + 𝛾: 0.0007011536816900648,
N14 + 1.5 He4 ⟶ Ne20 + 𝛾: 0.08158832557453448}
