Solar Composition#
Here we show how to work with the LoddersComposition class, which initializes a composition from the present day solar abundances of Lodders [2020]. To access the data tables, follow this link. This class gives the option to set a desired metallicity Z=0.0 and scales the Lodders abundances accordingly. If no metallicity is provided, it will be calculated from the Lodders data as (1 - H - He).
import numpy as np
import matplotlib.pyplot as plt
from ipywidgets import interact, FloatSlider
from pynucastro import networks
from pynucastro.nucdata import Nucleus
Setting solar composition and finding the top 10 most abundant isotopes#
def top_10_isotopes(comp, n=10):
fracs = sorted(comp.items(), key=lambda frac: frac[1], reverse=True)
return fracs[:n]
solar = networks.LoddersComposition() ## Default metallicity
top = top_10_isotopes(solar, n=10)
for nuc, X in top:
print(f"{nuc.short_spec_name:>6} X= {X:.4f}")
h1 X= 0.7462
he4 X= 0.2388
o16 X= 0.0064
c12 X= 0.0026
ne20 X= 0.0019
fe56 X= 0.0011
n14 X= 0.0007
si28 X= 0.0006
mg24 X= 0.0005
s32 X= 0.0003
Binning Lodders composition to a desired network#
If we wish to use a network that does not contain all isotopes that are in the Lodders composition, then we use Composition.bin_as which will bin the Lodders data to the desired network:
solar = networks.LoddersComposition()
new_comp = [Nucleus("p"),
Nucleus("he4"),
Nucleus("o16"),
Nucleus("c12"),
Nucleus("ne20"),
Nucleus("fe56"),
Nucleus("n14"),
Nucleus("si28"),
Nucleus("mg24"),
Nucleus("s32")] ## Desired network
binned_comp = solar.bin_as(new_comp)
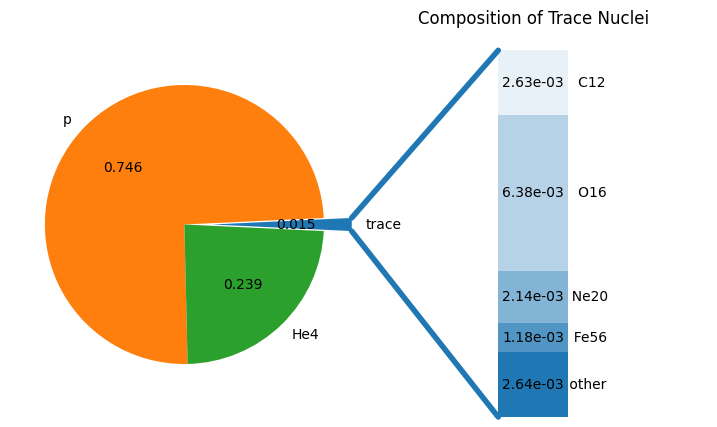
print("Sum of binned comp:", sum(binned_comp.values()))
fig = binned_comp.plot()
Sum of binned comp: 1.000000000000001
Scaling H and He with desired metallicty Z#
def xyz_from_comp(comp):
X = 0.0
Y = 0.0
Z = 0.0
for nuc, X_i in comp.items():
if nuc.Z == 1: # Total H
X += X_i
elif nuc.Z == 2: # Total He
Y += X_i
else:
Z += X_i # Total metals
return X, Y, Z
solar = networks.LoddersComposition()
X_solar, Y_solar, Z_solar = xyz_from_comp(solar)
print("Default solar metalicity is Z = ", Z_solar)
def plot_lodders(Z):
comp = networks.LoddersComposition(Z=Z)
X, Y , Z = xyz_from_comp(comp)
plt.figure(figsize=(4,4))
labels = ["H", "He", "Metals"]
values = [X,Y,Z]
x = np.arange(len(labels))
fig, ax = plt.subplots()
ax.bar(x, values)
ax.set_xticks(x)
ax.set_xticklabels(labels)
ax.set_ylabel("Mass Fractions")
ax.set_ylim(0, 1.0)
ax.set_title(f"Lodders Composition at Z = {Z:.3f}")
plt.show()
interact(
plot_lodders,
Z=FloatSlider(
value=Z_solar,
min=0.01,
max=0.04,
step=0.001,
description="Z",
readout_format=".3f"
)
)
Default solar metalicity is Z = 0.014964158698946859
<function __main__.plot_lodders(Z)>
